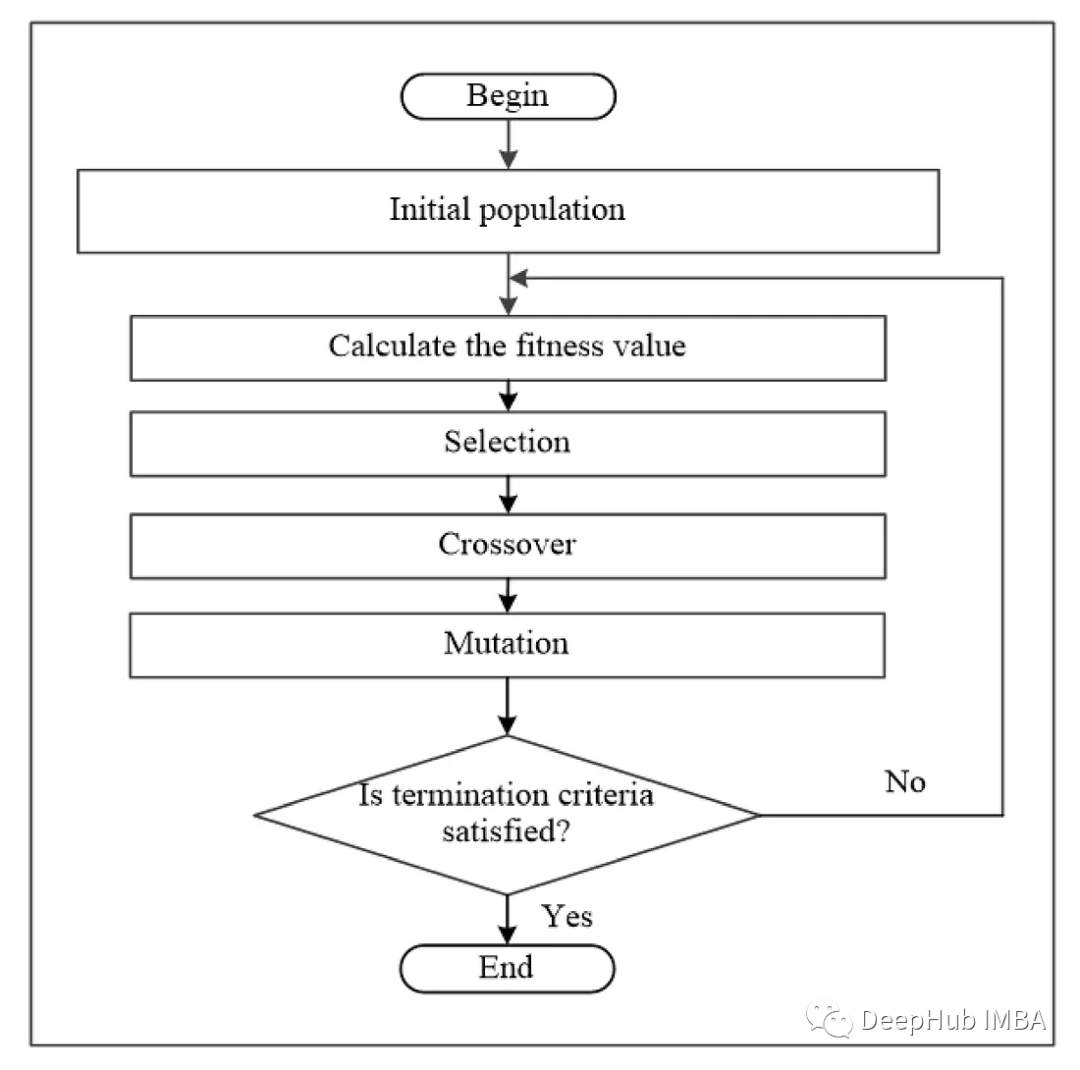
遗传算法是一种基于达尔文进化论的搜索启发式算法。该算法模拟了基于种群中最适合个体的自然选择。
遗传算法需要两个参数,即种群和适应度函数。根据适应度值在群体中选择最适合的个体。最健康的个体通过交叉和突变技术产生后代,创造一个新的、更好的种群。这个过程重复几代,直到得到最好的解决方案。
要解决的问题
本文中我们将使用遗传算法在迷宫中找到最短路径。
本文的的路径规划基于论文Autonomous Robot Navigation Using Genetic Algorithm with an Efficient Genotype Structure。
本文的代码基于两个假设:
1、代理不能对角线移动,也就是说只能左右,上下移动,不能走斜线。
2、迷宫的地图是已知的,但是代理不知道。
基因型
在由 N 列建模的导航环境中,路径可以由具有 N 个基因的基因型表示。 每个基因都是一个代表检查点坐标的元组。
所以我们的基因型如下,列式结构:
在列式结构中,我们假设每个基因都只放在一列中,例如,取一条大小为 8 的染色体,[(1,1), (4,2), (4,3), (6,4), (2,5), (3,6), (7,7), (8,8)]。 它会像这样放置:
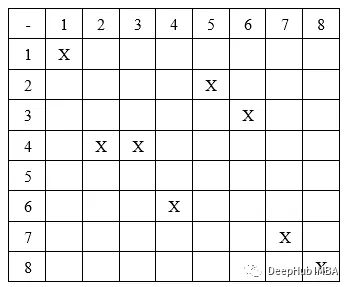
这里假设起始坐标是(1,1),目标坐标是(8,8)
有两种方法可以从一个检查点移动到另一个检查点:
先排成一行移动 或 先排成一列移动
行移动:先在一行中移动,然后在列中移动。 如果我们从前面的例子中获取染色体,我们得到的路径是:
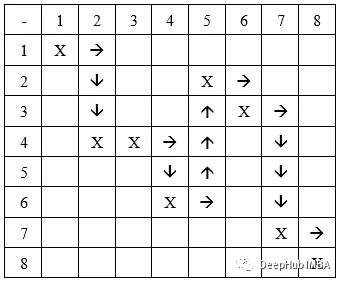
列移动:类似的,先在一列中移动,然后在一行中移动。 我们得到的路径是:
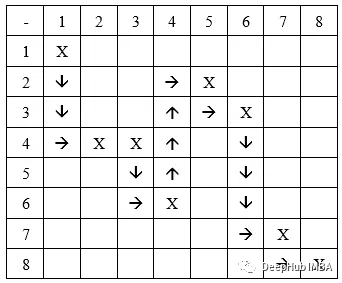
虽然这种基因型结构很好,但它不能为某些迷宫产生路径。 所以这种结构假定每个路径段都以连续的列结束。
实现遗传算法
本文使用python语言来实现遗传算法,并在最后有完整代码链接。
1、导入模块
这里的一个没见过的模块是pyamaze,它主要是python中生成迷宫。
fromrandomimportrandint, random, choice
frompyamazeimportmaze, agent
fromcopyimportdeepcopy, copy
importcsv2、定义常量和一些初始化
设置程序的不同参数并创建一个迷宫。
#initialize constants
POPULATIONSIZE=120
ROWS, COLS=16,8
#Set the mutation rate
MUTATION_RATE=100
# start and end points of the maze
START, END=1, COLS
#weights of path length, infeasible steps and number of turns
wl, ws, wt=2, 3, 2
#file name for storing fitness parameters
file_name='data.csv'
#initilize a maze object and create a maze
m=maze(ROWS, COLS)
m.CreateMaze(loopPercent=100)
maps=m.maze_map3、种群的创建
此函数创建种群并为每个个体随机生成方向属性。
defpopFiller(pop, direction):
"""
Takes in population and direction lists and fills them with random values within the range.
"""
foriinrange(POPULATIONSIZE):
# Generate a random path and add it to the population list
pop.append([(randint(1, ROWS), j) forjinrange(1, COLS+1)])
# Set the start and end points of the path
pop[i][0] = (START, START)
pop[i][-1] = (ROWS, END)
# Generate a random direction and add it to the direction list
direction.append(choice(["r", "c"]))
returnpop, direction4、路径计算
这一步使用了两个函数:inter_steps函数以两个坐标作为元组,例如(x, y)和方向信息来生成这些点之间的中间步数。path函数使用inter_steps函数通过循环每个个体的基因来生成它的路径。
definter_steps(point1, point2, direction):
"""
Takes in two points and the direction of the path between them and returns the intermediate steps between them.
"""
steps= []
ifdirection=="c": # column first
ifpoint1[0] point2[0]:
steps.extend([(i, point1[1]) foriinrange(point1[0] -1, point2[0] -1, -1)])
steps.append(point2)
elifdirection=="r": # row first
ifpoint1[0] point2[0]:
steps.extend([(i, point1[1] +1) foriinrange(point1[0], point2[0] -1, -1)])
else:
steps.append(point2)
returnsteps
defpath(individual, direction):
"""
Takes in the population list and the direction list and returns the complete path using the inter_steps function.
"""
complete_path= [individual[0]]
foriinrange(len(individual) -1):
# Get the intermediate steps between the current and the next point in the population
complete_path.extend(inter_steps(individual[i], individual[i+1], direction))
returncomplete_path 5、适应度评估
这个步骤使用两个函数来计算个体的适应度:pathParameters函数计算个体转弯次数和不可行步数,fitCal函数利用这些信息计算每个个体的适应度。 适应度计算的公式如下:

这些公式分别归一化了转身、不可行步长和路径长度的适应度。整体适应度计算公式如下所示:
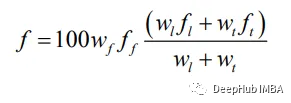
wf, wl, wt是定义参数权重的常数,分别是不可行的步数,路径长度和转弯次数。这些常量之前已经初始化。
fitCal函数有一个额外的关键字参数,即createCSV,它用于将不同的参数写入CSV文件。
defpathParameters(individual, complete_path, map_data):
"""
Takes in an individual, it's complete path and the map data
and returns the number of turns and the number of infeasible steps.
"""
# Count the number of turns in the individual's path
turns=sum(
1
foriinrange(len(individual) -1)
ifindividual[i][0] !=individual[i+1][0]
)
# Count the number of Infeasible steps in the individual's path
infeas=sum(
any(
(
map_data[complete_path[i]]["E"] ==0andcomplete_path[i][1] ==complete_path[i+1][1] -1,
map_data[complete_path[i]]["W"] ==0andcomplete_path[i][1] ==complete_path[i+1][1] +1,
map_data[complete_path[i]]["S"] ==0andcomplete_path[i][0] ==complete_path[i+1][0] -1,
map_data[complete_path[i]]["N"] ==0andcomplete_path[i][0] ==complete_path[i+1][0] +1,
)
)
foriinrange(len(complete_path) -1)
)
returnturns, infeas
deffitCal(population, direction, solutions,createCSV=True):
"""
Takes in the population list and the direction list and
returns the fitness list of the population and the solution found(infeasible steps equal to zero).
Args:
- population (list): a list of individuals in the population
- direction (list): a list of the direction for each individual
- solutions (list): a list of the solutions found so far
- createCSV (bool): a flag indicating whether to create a CSV file or not (default: True)
"""
# Initialize empty lists for turns, infeasible steps, and path length
turns= []
infeas_steps= []
length= []
# A function for calculating the normalized fitness value
defcalc(curr, maxi, mini):
ifmaxi==mini:
return0
else:
return1- ((curr-mini) / (maxi-mini))
# Iterate over each individual in the population
fori, individualinenumerate(population):
# Generate the complete path of individual
p=path(individual, direction[i])
# Calculate the number of turns and infeasible steps in the individual's path
t, s=pathParameters(individual, p, maps)
# If the individual has zero infeasible steps, add it to the solutions list
ifs==0:
solutions.append([deepcopy(individual), copy(direction[i])])
# Add the individual's number of turns, infeasible steps, and path length to their respective lists
turns.append(t)
infeas_steps.append(s)
length.append(len(p))
# Calculate the maximum and minimum values for turns, infeasible steps, and path length
max_t, min_t=max(turns), min(turns)
max_s, min_s=max(infeas_steps), min(infeas_steps)
max_l, min_l=max(length), min(length)
# Calculate the normalized fitness values for turns, infeasible steps, and path length
fs= [calc(infeas_steps[i], max_s, min_s) foriinrange(len(population))]
fl= [calc(length[i],max_l, min_l) foriinrange(len(population))]
ft= [calc(turns[i], max_t, min_t) foriinrange(len(population))]
# Calculate the fitness scores for each individual in the population
fitness= [
(100*ws*fs[i]) * ((wl*fl[i] +wt*ft[i]) / (wl+wt))
foriinrange(POPULATIONSIZE)
]
# If createCSV flag is True, write the parameters of fitness to a CSV file
ifcreateCSV:
withopen(file_name, 'a+', newline='') asf:
writer=csv.DictWriter(f, fieldnames=['Path Length', 'Turns', 'Infeasible Steps', 'Fitness'])
writer.writerow({'Path Length': min_l, 'Turns': min_t, 'Infeasible Steps': min_s, 'Fitness': max(fitness)})
returnfitness, solutions6、选择
这个函数根据适应度值对总体进行排序。
defrankSel(population, fitness, direction):
"""
Takes in the population, fitness and direction lists and returns the population and direction lists after rank selection.
"""
# Pair each fitness value with its corresponding individual and direction
pairs=zip(fitness, population, direction)
# Sort pairs in descending order based on fitness value
sorted_pair=sorted(pairs, key=lambdax: x[0], reverse=True)
# Unzip the sorted pairs into separate lists
_, population, direction=zip(*sorted_pair)
returnlist(population), list(direction)7、交叉
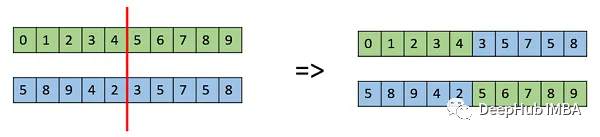
这个函数实现了单点交叉。它随机选择一个交叉点,并使用种群人口的前一半(父母)来创建后一半,具体方法如下图所示:
defcrossover(population, direction):
"""
Takes in the population and direction lists and returns the population and direction lists after single point crossover.
"""
# Choose a random crossover point between the second and second-to-last gene
crossover_point=randint(2, len(population[0]) -2)
# Calculate the number of parents to mate (half the population size)
no_of_parents=POPULATIONSIZE//2
foriinrange(no_of_parents-1):
# Create offspring by combining the genes of two parents up to the crossover point
population[i+no_of_parents] = (
population[i][:crossover_point] +population[i+1][crossover_point:]
)
# Choose a random direction for the offspring
direction[i+no_of_parents] =choice(["r", "c"])
returnpopulation, direction8、变异
通过将基因(即tuple (x, y))的x值更改为范围内的任意数字来实现插入突变。元组的y值保持不变,因为我们假设迷宫中的每一列都应该只有一个检查点。
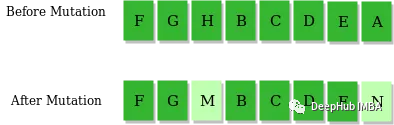
有几个参数可以调整,mutation_rate和no_of_genes_to_mutate。
defmutation(population, mutation_rate, direction, no_of_genes_to_mutate=1):
"""
Takes in the population, mutation rate, direction and number of genes to mutate
and returns the population and direction lists after mutation.
"""
# Validate the number of genes to mutate
ifno_of_genes_to_mutateCOLS:
raiseValueError(
"Number of genes to mutate must not be greater than number of columns"
)
foriinrange(POPULATIONSIZE):
# Check if the individual will be mutated based on the mutation rate
ifrandom() 9、最佳解
该函数根据解的路径长度返回最佳解。
defbest_solution(solutions):
"""Takes a list of solutions and returns the best individual(list) and direction"""
# Initialize the best individual and direction as the first solution in the list
best_individual, best_direction=solutions[0]
# Calculate the length of the path for the best solution
min_length=len(path(best_individual, best_direction))
forindividual, directioninsolutions[1:]:
# Calculate the length of the path for the best solution
current_length=len(path(individual, direction))
# If the current solution is better than the best solution, update the best solution
ifcurrent_length10、整合
整个算法的工作流程就像我们开头显示的流程图一样,下面是代码:
defmain():
# Initialize population, direction, and solution lists
pop, direc, sol= [], [], []
# Set the generation count and the maximum number of generations
gen, maxGen=0, 2000
# Set the number of solutions to be found
sol_count=1
# Set the number of solutions to be found
pop, direc=popFiller(pop, direc)
# Create a new CSV file and write header
withopen(file_name, 'w', newline='') asf:
writer=csv.DictWriter(f, fieldnames=['Path Length', 'Turns', 'Infeasible Steps', 'Fitness'])
writer.writeheader()
# Start the loop for the genetic algorithm
print('Running...')
whileTrue:
gen+=1
# Calculate the fitness values for the population and identify any solutions
fitness, sol=fitCal(pop, direc, sol, createCSV=True)
# Select the parents for the next generation using rank selection
pop, direc=rankSel(pop, fitness, direc)
# Create the offspring for the next generation using crossover
pop, direc=crossover(pop, direc)
# mutate the offsprings using mutation function
pop, direc=mutation(pop, MUTATION_RATE, direc, no_of_genes_to_mutate=1)
# Check if the required number of solutions have been found
iflen(sol) ==sol_count:
print("Solution found!")
break
# Check if the maximum number of generations have been reached
ifgen>=maxGen:
print("Solution not found!")
flag=input("Do you want to create another population(y/n): ")
# if flag is 'y', clear the population and direction lists and refill them
ifflag=='y':
pop, direc= [], []
pop, direc=popFiller(pop, direc)
gen=0
continue
# If flag is 'n' exit the program
else:
print("Good Bye")
returnNone
# Find the best solution and its direction
solIndiv, solDir=best_solution(sol)
# Generate the final solution path and reverse it
solPath=path(solIndiv, solDir)
solPath.reverse()
# Create an agent, set its properties, and trace its path through the maze
a=agent(m, shape="square", filled=False, footprints=True)
m.tracePath({a: solPath}, delay=100)
m.run()
if__name__=="__main__":
# Call the main function
main()可视化和结果展示
最后为了能看到算法的过程,可以可视化不同参数随代数的变化趋势,使用matplotlib和pandas绘制曲线。

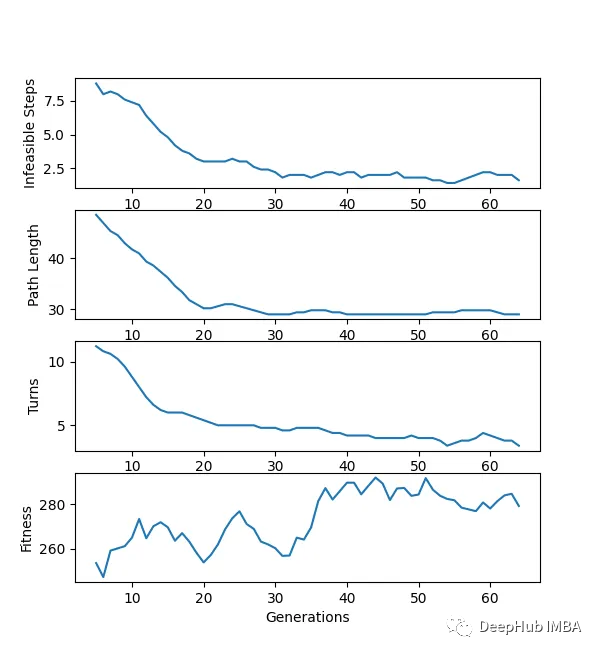
下面是一个使用“loopPercent = 100”的15 * 15迷宫的结果:
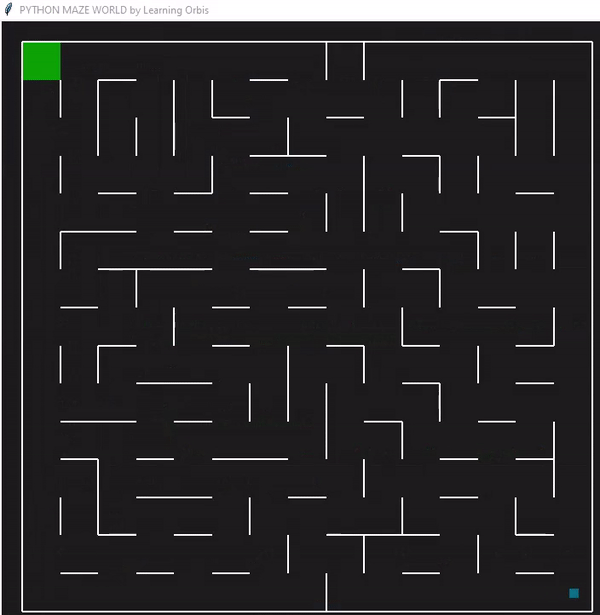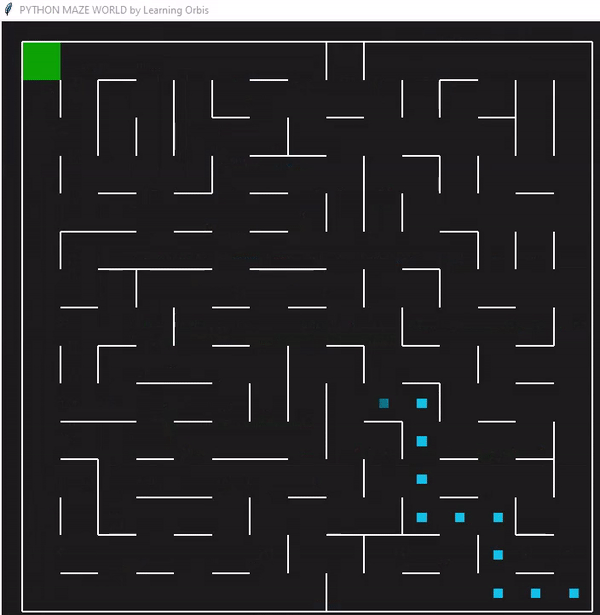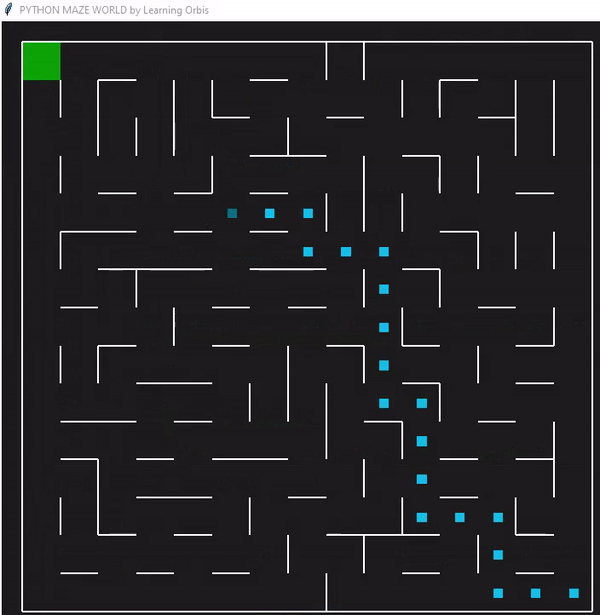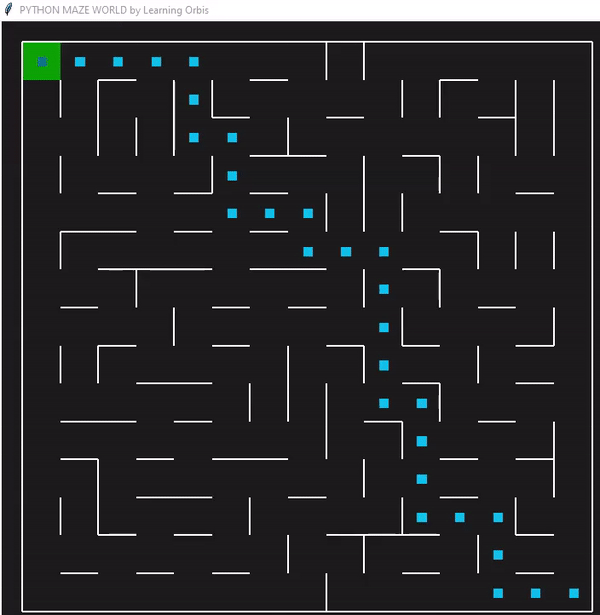
我们可以对他进行一些简单的分析:下图是我结果中得到的趋势线。“不可行的步数”显著减少,“路径长度”和“转弯数”也显著减少。经过几代之后,路径长度和转弯数变得稳定,表明算法已经收敛到一个解决方案。
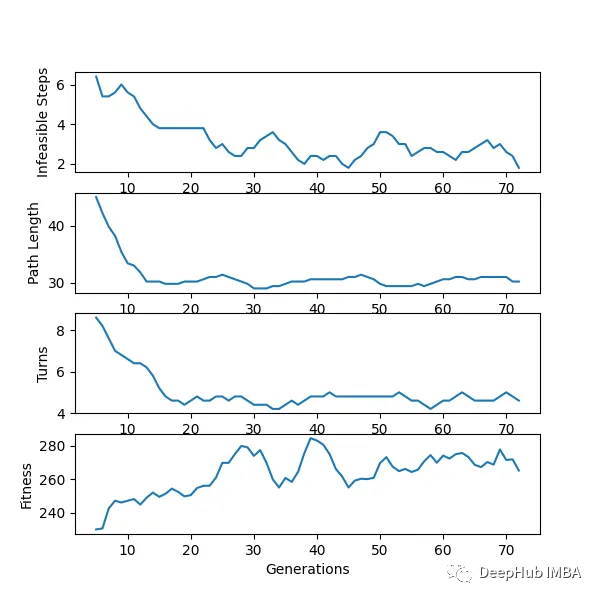
适应度曲线不一致的可能原因:
当一个个体在种群中的适应度最大时,不意味着它就是解决方案。但它继续产生越来越多的可能解,如果一个群体包含相似的个体,整体的适合度会降低。
下面一个是是使用“loopPercent = 100”的10 * 20迷宫的结果:
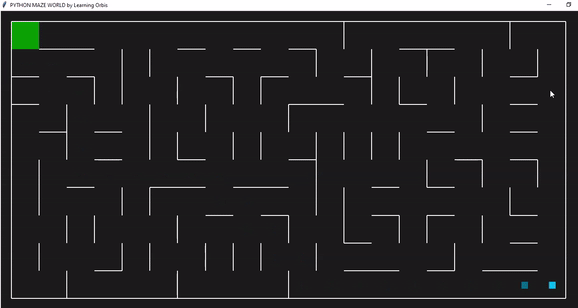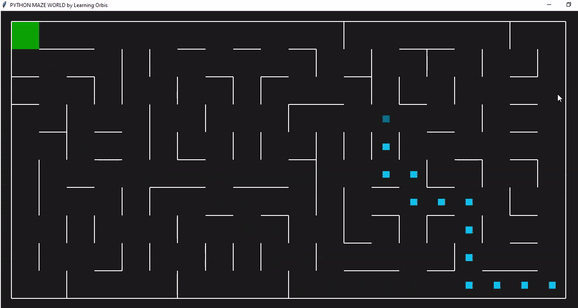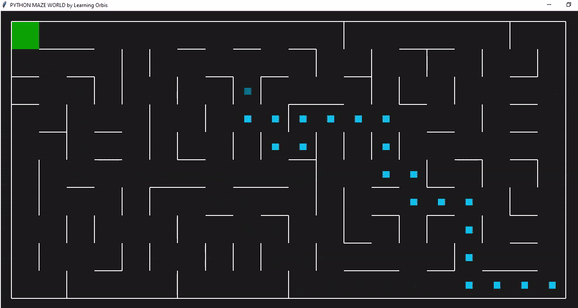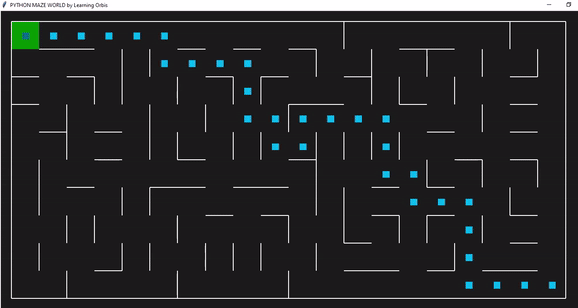
趋势线与之前的迷宫相似:
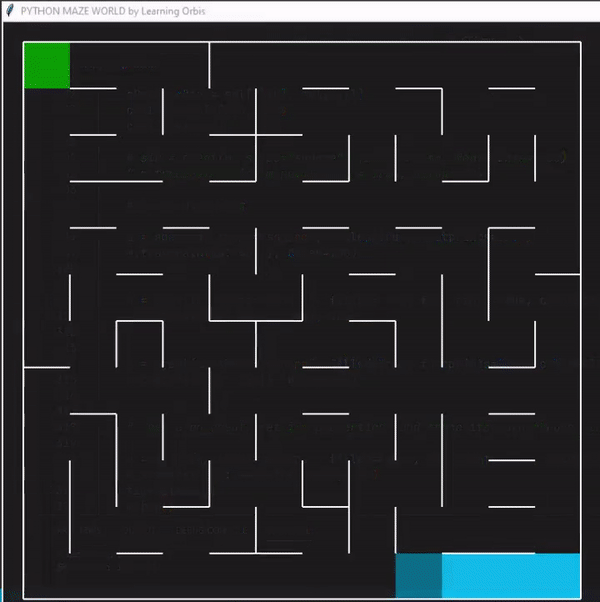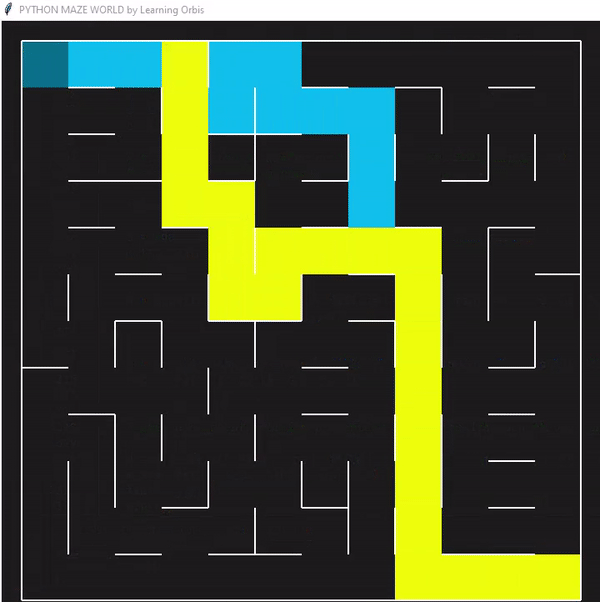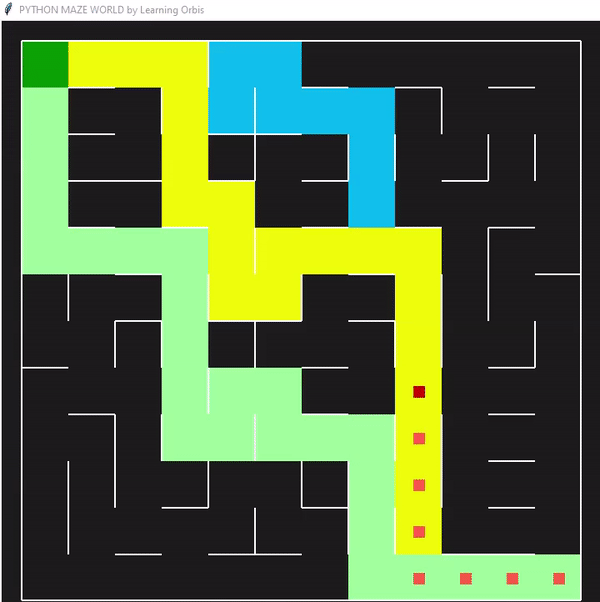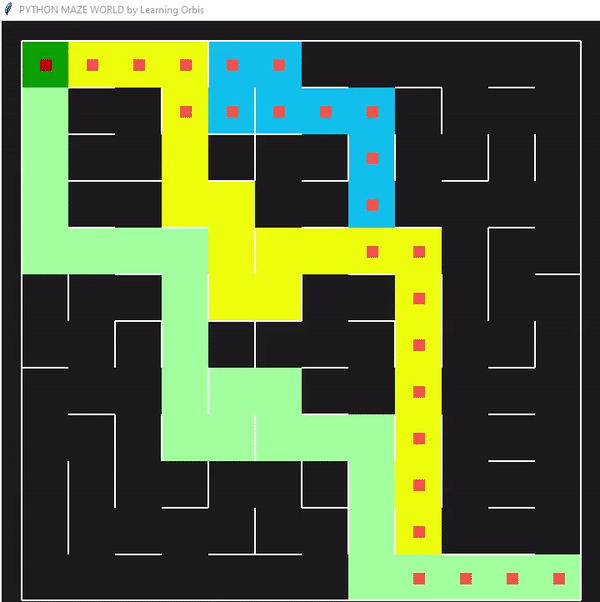
使用“loopPercent = 100”的12 × 12迷宫的结果:
程序运行后找到的三个解决方案,并从中选择最佳解决方案。红色的是最佳方案。最佳解决方案是根据路径长度等标准选择的。与其他解决方案相比,红色代理能够找到通过迷宫的有效路径。这些结果证明了该方案的有效性。
一些数据指标的对比
计算了10个不同大小的迷宫的解决方案所需时间的数据。
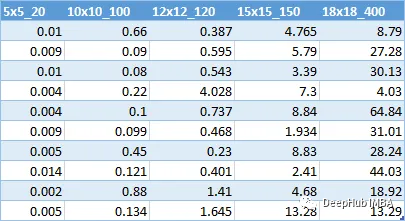
随着迷宫规模的增加,时间几乎呈指数增长。这意味着用这种算法解决更大的迷宫是很有挑战性的。
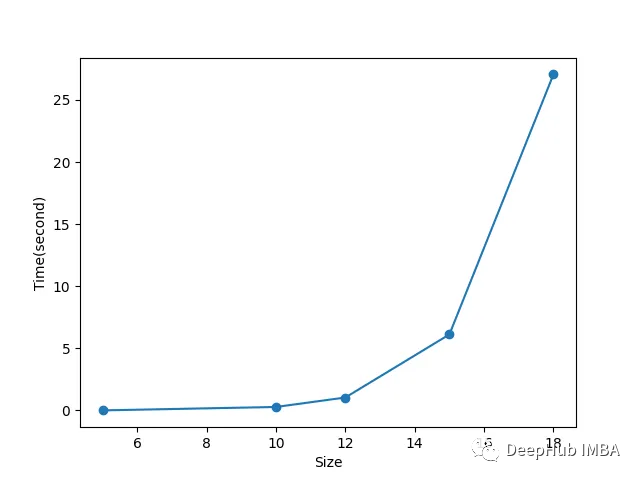
这是肯定的:
因为遗传算法是模拟的自然选择,有一定的随机性,所以计算量很大,特别是对于大而复杂的问题。我们选择的实现方法也适合于小型和简单的迷宫,基因型结构不适合大型和复杂的迷宫。迷宫的结果也取决于初始总体,如果初始总体是好的,它会更快地收敛到解决方案,否则就有可能陷入局部最优。遗传算法也不能找到最优解。
总结
本文我们成功地构建并测试了一个解决迷宫的遗传算法实现。对结果的分析表明,该算法能够在合理的时间内找到最优解,但随着迷宫大小的增加或loopPercent的减少,这种实现就会变得困难。参数POPULATION_SIZE和mutation_rate,对算法的性能有重大影响。本文只是对遗传算法原理的一个讲解,可以帮助你了解遗传算法的原理,对于小型数据可以实验,但是在大型数据的生产环境,请先进行优化。
https://avoid.overfit.cn/post/db9f6af37de141c99d0a054ff767dc71
作者:Dayan Hafeez
服务器托管,北京服务器托管,服务器租用 http://www.fwqtg.net

